How ProteoCombiner works

This Nature Protocol Exchange explains how ProteoCombiner works and how the parameters must be used. There are many details that need to be read carefully.
Get started
After installing ProteoCombiner, the user is able to combine data from different proteomics approaches.
1. Follow the step by step below to load all the necessary files and perform the first data combination.
- In Supplementary files, download the protein databases used in bottom-up and top-down proteomics searches.
- Also in Supplementary files, download the result files obtained in bottom-up (MaxQuant: peptides.txt, proteinGroups.txt and msms.txt, and PatternLab for Proteomics: ProteoCombinerResults.sepr) and top-down (20170105_L_MaD_ColC4_Ecoli20161108-01-02-03_02_PSMs.txt) proteomics searches.
- If you want to visualize the identified spectra, download the RAW files at PRIDE Repository, which have been used in the manuscript.
- Specify the directory where are the bottom-up files: protein database (in our example, a FASTA file was used) and all bottom-up search engine(s) result files (in our example, MaxQuant & PatternLab for Proteomics result files). If you have been done the step 3, the following RAW files need to be in the same directory: 20161222_Q1_MD_colQ1-33_Ecoli_Mat_B1bis.raw, 20161222_Q1_MD_colQ1-33_Ecoli_Mat_B2.raw and 20161222_Q1_MD_colQ1-33_Ecoli_Mat_B3.raw.
- Specify the directory where are the top-down files: protein database (in our example, an XML file was used) and all top-down search engine(s) result files (in our example, ProSight result file). If you have been done the step 3, the following RAW files need to be in the same directory: 20170105_L_MaD_ColC4_Ecoli20161108-01_01.raw, 20170105_L_MaD_ColC4_Ecoli20161108-02_01.raw and 20170105_L_MaD_ColC4_Ecoli20161108-03_01.raw.
- By clicking on GO button, the combination will start.
2. Follow the step by step below to load a data combination performed as example.
- In Supplementary files, download the ProteoCombiner output file (*.pcmb). PS: Uncompress proteocombiner-output.zip for obtaining the *.pcmb file.
- Load the ProteCombiner result, by clicking on File menu → Load Results (or press CTRL + O) and select the results.pcmb downloaded from step 1.
- A new window is open.
- The result file (*.pcmb) can be open just by double-clicking on it.
Note: The images below summarize how ProteoCombiner works.
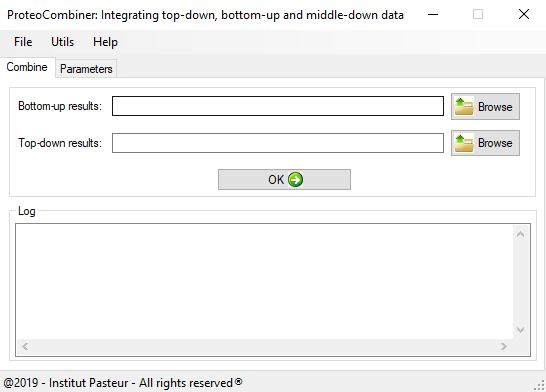
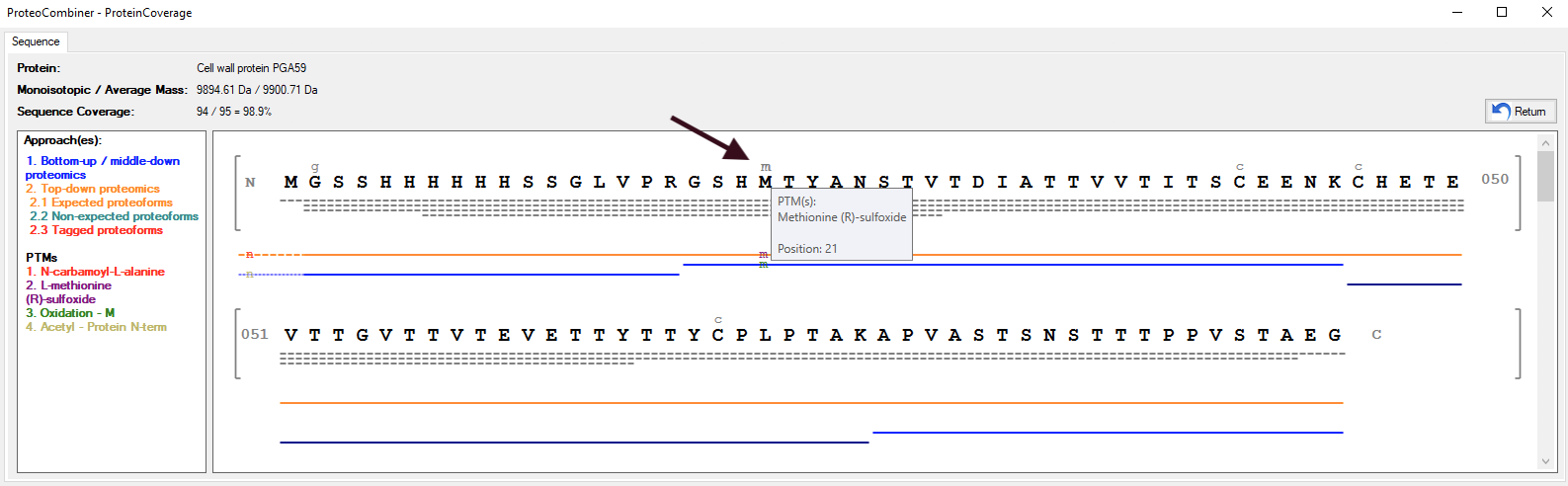
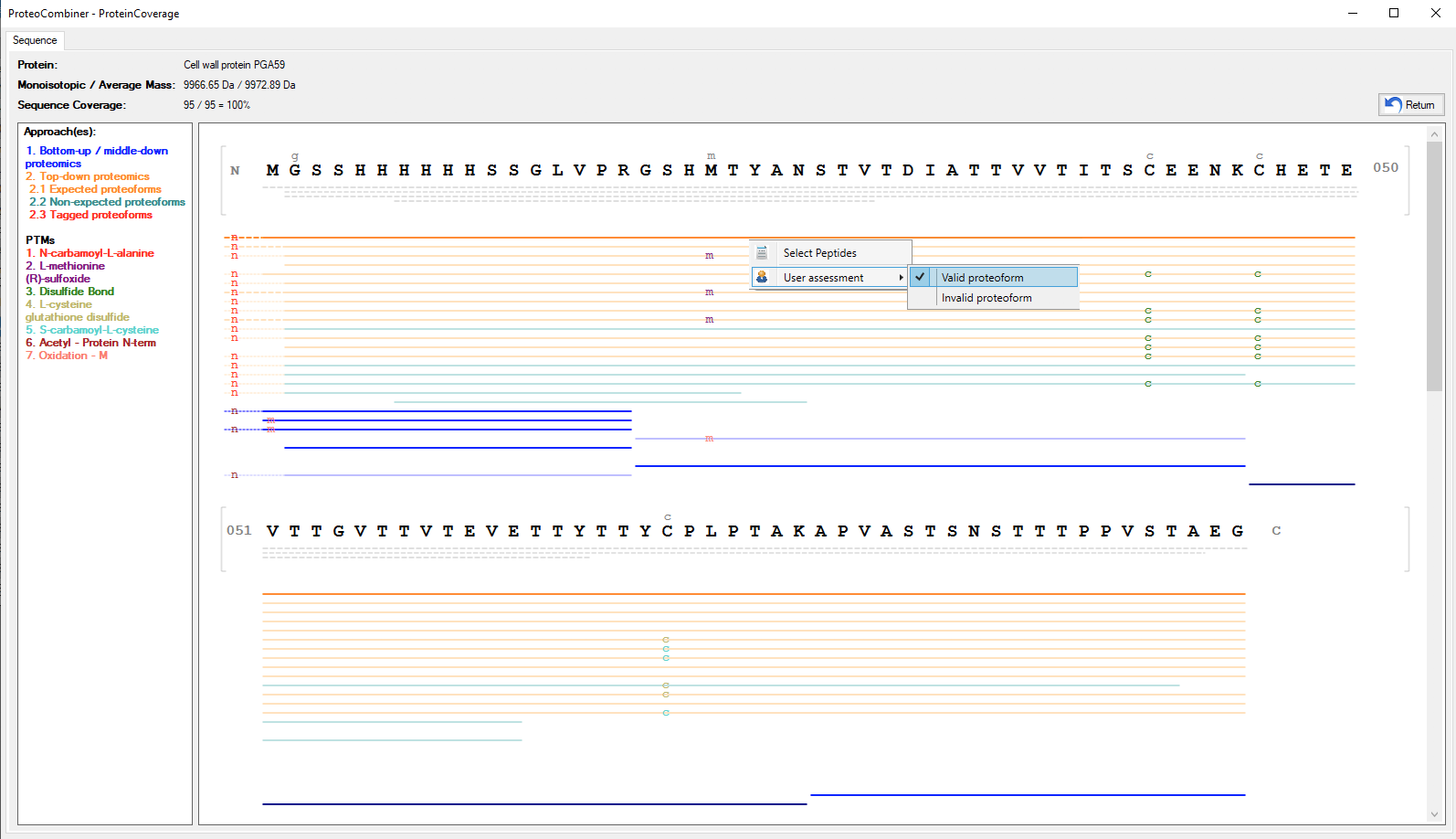
Figure 1 shows the main Graphical User Interface of ProteoCombiner. It provides an unmatched toolbox for rapid and easy visualization, manual validation and comparison of the identified proteoform sequences, including post-translational modifications (PTM) characterization (Figure 2a and Figure 2b).
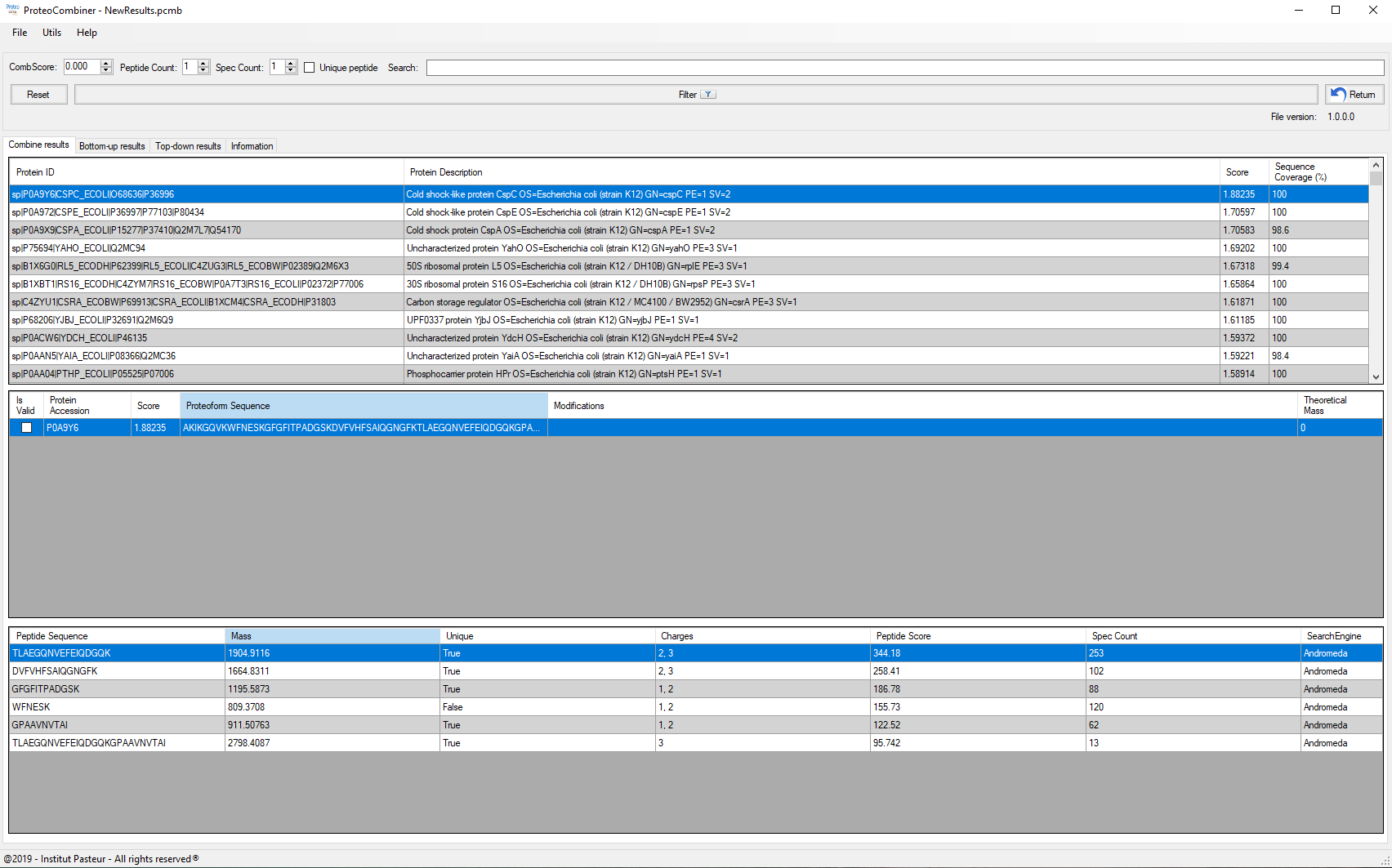
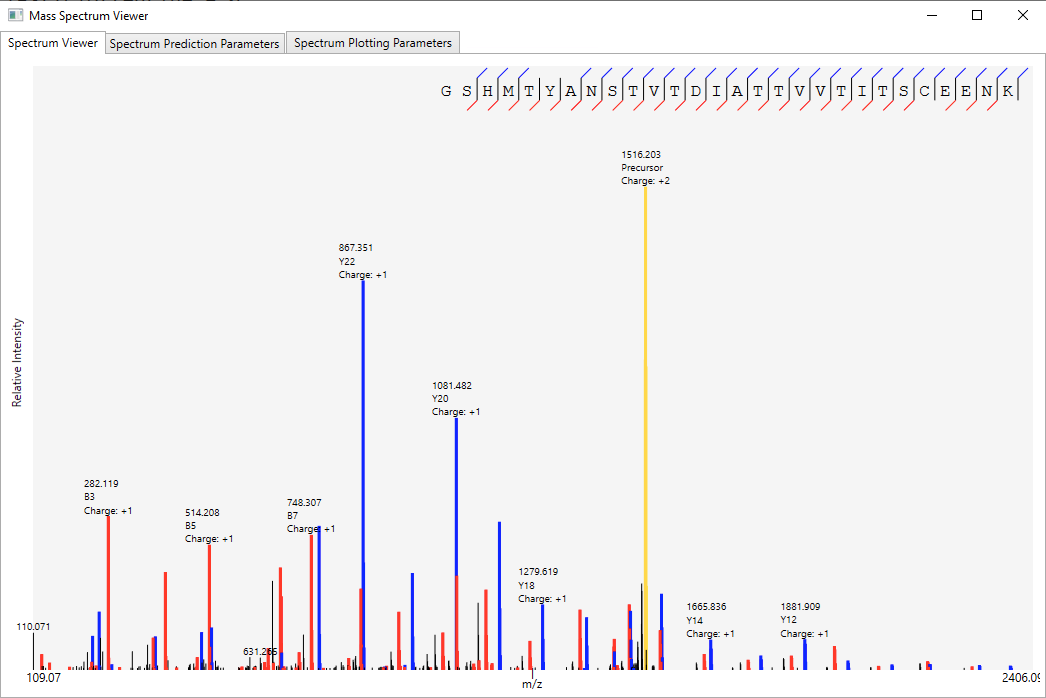
Once data combination is done, a dynamic report is generated (Figure 3), and on Combine Results tab allows the sorting/searching of results according to a user-specified criterion, as well as accessing the spectrum viewer (Figure 4) by double-clicking on an peptide identification. The user can also access the Protein Coverage (Figures 2a and 2b) by double-clicking on an identified protein in the first table or on an identified proteoform / peptide in the following table.